
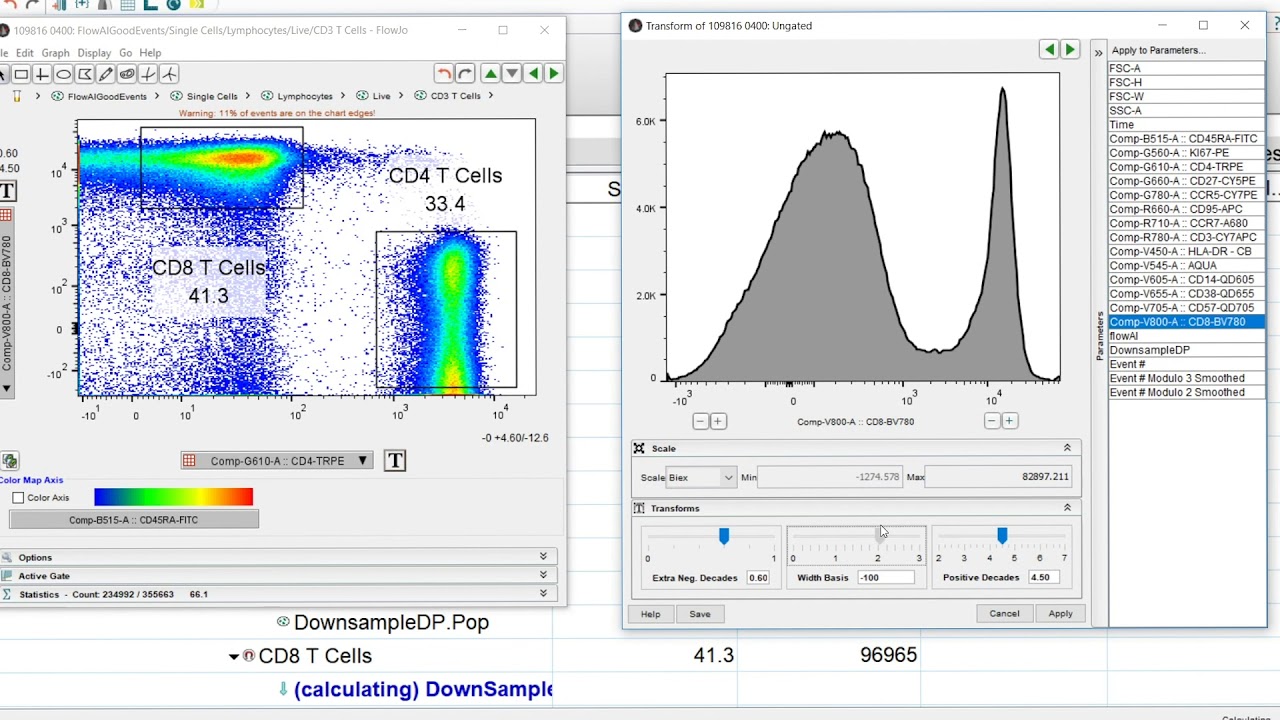
Is there a way to remove FlowJo's transformations from the exported FCS files? Alternatively, is there a way to select the portion of the data for export and then remove all transformations in FlowJo?ģ. These files' expression values differ when imported using transformation = "linearize", but the output of keyword($P¡n❾) looks the same for both files.Ģ. Does transformation = "linearize" in read.FCS only act on data that are stored with different exponentiation? I've received some compensated and compensated + transformed files that are identical when read in with transformation = F. Clearly, we do not want to double-transform any of the data.ġ. When the analysts receive the data, we generally want one transformation (typically either logicle or asinh with b=1/150) applied to each of the markers. The exported FCS files then often end up with some markers being transformed (sometimes this affects the most recently opened marker alone, sometimes this affects nearly all of the markers). The biexponential transformation is applied to some of the data when our wet-lab folks open and start gating the data. I'm a relative novice when it comes to flow data, and I have a couple of general questions about shifting the data from FlowJo to R and performing the pre-processing steps.

This data is then delivered to our computational group for additional analysis. Scientists that perform the benchwork and acquisition of flow cytometry data at my institute export FCS files from FlowJo after both compensating the data and removing dead cells/populations that are not of interest.


 0 kommentar(er)
0 kommentar(er)
